Overview
Single Cell Spatial View grant application resources10x Genomics provides instruments, consumables and software to measure biology at large scale and unprecedented resolution. With these three complementary platforms – Chromium, Visium, and Xenium – 10x Genomics offers unparalleled flexibility and precision.
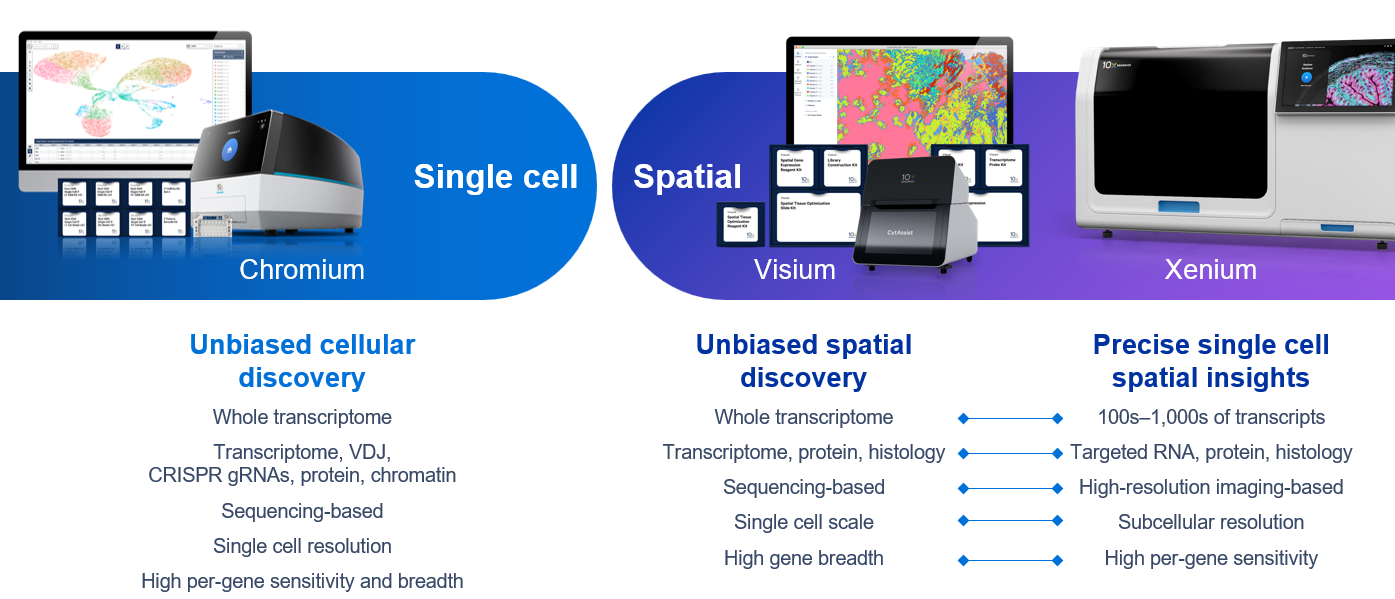
- Chromium Single Cell: Unbiased whole transcriptome results for dissociated samples, up to millions of cells.
- Visium Spatial: Unbiased whole transcriptome results for tissue sections with spatial context and single cell–scale resolution.
- Xenium In Situ: Targeted results for up to 5,000 genes with subcellular spatial context.
Low per-sample costs for single cell studies

Easy on-chip multiplexing for 3′ and 5′ scRNA-seq provides results you can trust—no extra steps, no extra costs. At every stage of your research, accurate, reliable performance is critical. So is your budget. 10x Genomics’ flagship gene expression (3′) and immune profiling (5′) assays—now available under the new product family names GEM-X Universal 3′ Gene Expression and GEM-X Universal 5′ Gene Expression—enable you to meet those goals, with results you can trust at more cost-effective price points.
New on-chip multiplexing for these assays lets you go even further, with a simple workflow you’ll love. Confidently run cost-effective single cell studies with limited samples. Up to 70% cell capture rates, unmatched sensitivity, and easy multiplexing at low per-sample costs. With GEM-X Universal 3’ and 5’ Gene Expression Multiplex, it all adds up to higher performance at small-scale prices. This unique technology is now available for pre-order and will be shipping before the end of 2024.
Lower cost per sample for high-quality single cell

- Perform reliable, high-quality single cell studies at low per-sample costs
- Run up to 8 samples and up to 40K cells per chip at lower costs with reduced reagent volumes and the ability to add replicates without increasing your spend
- Get the highest return on your sequencing budget with unmatched gene sensitivity
- Uncover deeper insights with multiomic capabilities, including protein detection, TCR/BCR profiling, CRISPR screens, and more
Easy on-chip multiplexing—no extra steps
- Batch 4 independent samples per set and collect all partitioned cells in the same recovery well—no extra steps required
- Expand multiplexing to more samples with a species-agnostic approach that does not require sample tagging upstream of chip loading
- Ease technical restrictions with lower cell inputs for multiplexing limited samples like stem cells or flow-sorted cells for rare disease studies
- Conduct more statistically powerful studies by enabling biological or technical replicates without increasing costs
Exceptional GEM-X performance for Universal 3’ and 5’ Gene Expression Multiplex 
- Count on the same industry-leading performance demonstrated with the standard workflows for GEM-X Universal 3’ and 5’ assays
- Make the most of every run with up to 70% cell capture and high gene sensitivity for multiplexed samples
- Generate highly reproducible results across runs, users, and timepoints
- Ensure your success with robust, optimised protocols and automated cell partitioning that removes the risk of manual error inherent in time-consuming instrument-less workflows
Performance gains. Expanded scale. New low prices.

Scale to millions of cells for low per-cell cost

- Conduct large-scale studies with a 2x increase in throughput, easily scalable to millions of cells and hundreds of samples
- Profile 25K cells per sample and 320K cells per channel with in-line multiplexing
- Run up to 2.56M cells per chip and choose kit options that achieve low per-cell costs
- Maximise discovery power for high-throughput screens, atlassing, multi-site translational studies, and large patient cohort studies
Unlock previously inaccessible samples

- Generate reliable, high-quality data from more samples, including fresh, frozen, and fixed samples—even FFPE
- Explore limited samples like small tissue biopsies and flow-sorted cells with new input recommendations as low as 25K cells per sample
- Ensure you get the most out of precious samples, with improved sample recovery up to 80%
- Achieve high performance on challenging and low-quality samples, with 3-fold probe coverage for 90% of genes
Flex offers ultimate flexibility in sample prep options with the ability to fix, batch, and run samples on your own schedule without compromising on data quality.



