Millennium Science
Enabling next-generation research
- Home
- Instruments
- Reagents
- Applications
- Latest Updates
- Promotions
- Events
- About us
- Contact Us
Chromium Single Cell
Visium Spatial
Instruments
Single cell technology is now better equipped to help unravel biological complexity. The latest addition to the Chromium platform, Single Cell Gene Expression Flex, brings our most sensitive single cell assay to your bench, with flexible options for everything from pilot experiments to million-cell studies.
This powerful probe-based assay boosts your discovery power, enabling you to:
Profile thousands of genes at the single cell level for unbiased characterization of cell types and cell states

Profile thousands of genes at the single cell level by barcoding mRNA at the 5' end, for unbiased characterisation of cell types and cell states
The first commercial single cell multiomics product for simultaneous profiling of gene expression and chromatin landscape at single cell resolution.

Capture multiple measurements simultaneously at the single cell level for deeper characterisation of cell types and states
Product Sheet: Simultaneous profiling of the transcriptome and epigenome from the same cell

Chromium Single Cell ATAC v2 is now shipping!
Chromium Single Cell Assay for Transposase Accessible Chromatin (ATAC) is a powerful method to understand gene regulation and characterise cell types and states, all at the single cell level. Building upon v1.1 chemistry, the v2 assay provides increased sensitivity and improved signal-to-noise ratio, enhancing your ability to detect rare peaks.
Product Sheet: Profiling chromatin accessibility at single cell resolution
Prep for success:
10x Genomics' new Chromium Nuclei Isolation Kit removes the complexities of nuclei isolation with an easy-to-use solution specifically designed for use with 10x Genomics single cell assays for gene expression and chromatin accessibility.
What sample types are compatible with the Chromium Nuclei Isolation Kit
10x Genomics 5' CRISPR is now available for order and shipment!
5' CRISPR Product Sheet | CRISPR Infographic | User Guide & Protocols
What is the difference between the 10x Genomics Single Cell 5' and 3' CRISPR Screening assays, and which should I use?
Our 5' CRISPR screening assay is compatible with most pre-existing Cas9 guide RNA vectors. If you have an existing CRISPR library or are starting a single cell CRISPR screen for the first time, we recommend using our Single Cell 5' v2 assay which is compatible with most guide libraries with little to no optimization required.
Our 3' CRISPR screening assay requires that a specific Capture Sequence be inserted into the vector at the appropriate location for capture of the guide. If you want to compare your single cell CRISPR experiment to data from previous experiments using 3' single cell gene expression with Feature Barcode technology, we recommend continuing to use our Single Cell 3' v3.1 assay to minimize batch effects.
A clear path to gene function with single cell CRISPR screening
Comprehensive antigen-specific clonotype discovery in one week
BEAM (Barcode Enabled Antigen Mapping) is a multiplexed antigen screening workflow that empowers rapid discovery of antigen-specific B-cell (BEAM-Ab) and T-cell (BEAM-T) clonotypes. Obtain comprehensive antigen-specific cellular profiles—including full-length paired V(D)J sequences, gene expression, and cell surface proteins at single cell resolution—to identify rare, antigen-reactive T and B cells for accelerating therapeutic discovery.

Transform antibody discovery with the fastest hit generation workflow, powered by multiplexed antigen screening against thousands of single B cells.
Antigen-specific T cells don't have to be hard to find anymore. Map antigens to T-cell receptors with off-the-shelf ease and unparalleled cellular characterisation.
Visualise gene expression within the tissue context. Available for FFPE and Fresh Frozen samples.

Fresh Frozen - Product Sheet FFPE - Product Sheet
Gain a new perspective on tissue complexity with simultaneous gene and protein spatial profiling
Whether you're just starting out with single cell, expanding your capabilities, or seeking an automated way to partition individual cells and generate barcoded, sequencing-ready libraries, you're covered. 10x Genomics' proven Next GEM technology provides a trusted path to advance your understanding of cell diversity and function.

 The most flexible way to single cell.
The most flexible way to single cell.
Expand your single cell capabilities with the most advanced hardware and highest throughput flexibility, enabling multiomic analysis of hundreds to millions of cells.
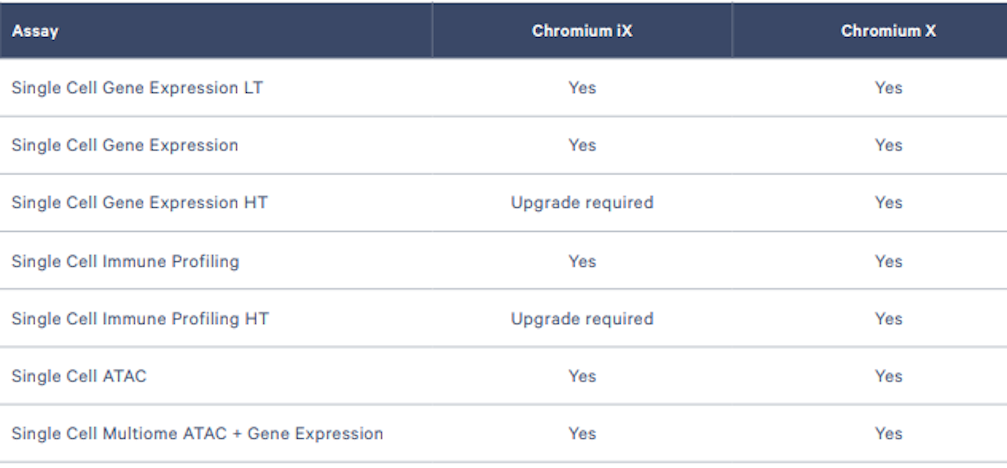
Perform multiomic analysis of hundreds to tens of thousands of single cells with this compact system.
Assay compatibility:
Combine single cell partitioning, barcoding, and/or library construction with standardised, automated workflows to maximize your productivity.
Seamlessly transition between standard histology workflows and analyte capture onto Visium slides for in-depth expression analysis.
 With Visium CytAssist you can:
With Visium CytAssist you can:
Workflow: Facilitate transfer of transcriptomic analytes in FFPE samples with Visium CytAssist
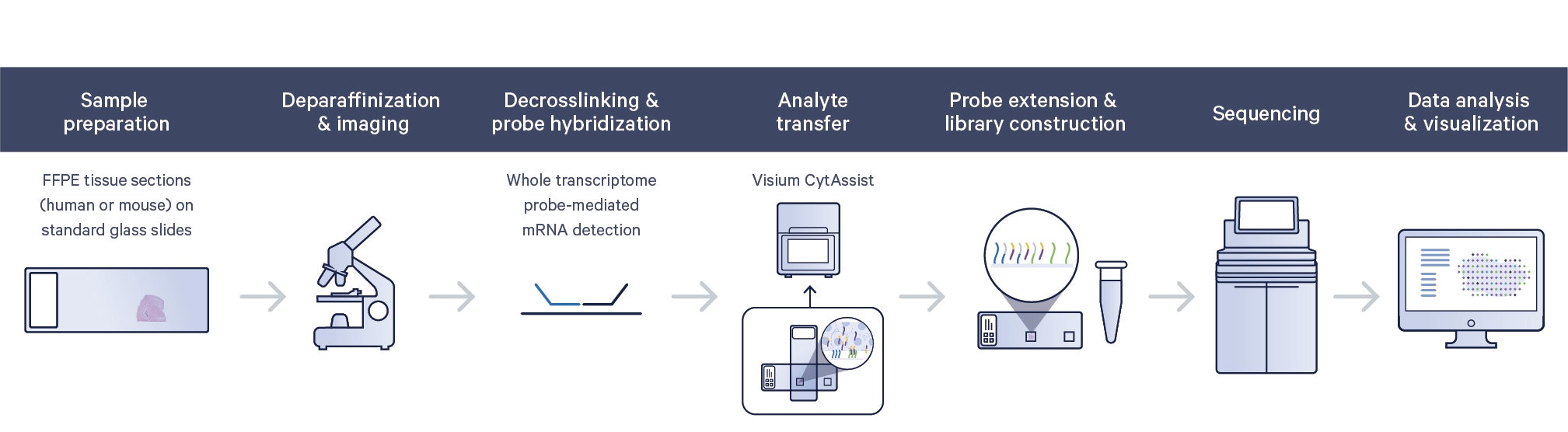
Build spatial transcriptome maps with Xenium In Situ
The Xenium workflow starts with sectioning tissues onto a microscope slide. The sections are then treated to access the RNA for labeling with circularizable DNA probes. Ligation of the probes then generates a circular DNA probe which is enzymatically amplified and bound with fluorescent oligos, creating bright, easy to image signal that has a high signal-to-noise ratio. The slide is then placed in the Xenium In Situ Analyzer where the sample then undergoes successive rounds of fluorescent probe hybridization, imaging, and removal. An optical signature specific to each gene is generated, enabling identification of the target gene. Finally, a spatial map of the transcripts is built across the entire tissue section.


This proprietary Next GEM technology fuels our Chromium System with an innovative reagent delivery system, set of algorithms and turnkey software analysis tools that enable the discovery of previously inaccessible genetic information at massive rate and scale.
The Next GEM technology is built on a new chip architecture that integrates seamlessly into existing solutions. This technology will enable future solutions and product improvements. The Next GEM technology combines new chips and reagents, and is currently offered for the following solutions:
Uncover the "where" for every "What".
The relationship between cells and their relative locations within tissue is critical to understanding normal development and disease pathology. Spatial transcriptomics is a groundbreaking molecular profiling method that allows scientists to measure all the gene activity in a tissue sample and map where the activity is occurring. Already this technology is leading to new discoveries that are proving instrumental in helping scientists gain a better understanding of biological processes and disease.
Available for: